I'm using hemoglobin for my example. Search Wikipedia for your protein (Hemoglobin). Notice that hemoglobin has 3 subunits. I am going to choose HBA1 for my study. I click on HBA1.
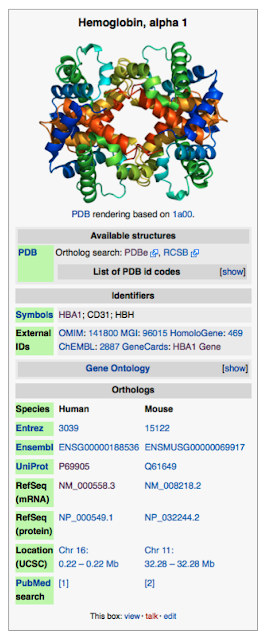 At right is part of the page for Hemoglobin subunit alpha
1. For the purposes of this exercise,
you want to compare mRNA sequences, so click on RefSeq(mRNA)
At right is part of the page for Hemoglobin subunit alpha
1. For the purposes of this exercise,
you want to compare mRNA sequences, so click on RefSeq(mRNA)
Scroll down the the bottom of the page that opens and you will find the DNA sequence corresponding to the mRNA as shown below for hemoglobin subunit a.
This is the mRNA sequence from a eukaryote. Which of the following does it contain?
- Promoter
- Start codon
- Stop codon
- Introns
- Exons
ORIGIN 1 actcttctgg tccccacaga ctcagagaga acccaccatg gtgctgtctc ctgccgacaa 61 gaccaacgtc aaggccgcct ggggtaaggt cggcgcgcac gctggcgagt atggtgcgga 121 ggccctggag aggatgttcc tgtccttccc caccaccaag acctacttcc cgcacttcga 181 cctgagccac ggctctgccc aggttaaggg ccacggcaag aaggtggccg acgcgctgac 241 caacgccgtg gcgcacgtgg acgacatgcc caacgcgctg tccgccctga gcgacctgca 301 cgcgcacaag cttcgggtgg acccggtcaa cttcaagctc ctaagccact gcctgctggt 361 gaccctggcc gcccacctcc ccgccgagtt cacccctgcg gtgcacgcct ccctggacaa 421 gttcctggct tctgtgagca ccgtgctgac ctccaaatac cgttaagctg gagcctcggt 481 ggccatgctt cttgcccctt gggcctcccc ccagcccctc ctccccttcc tgcacccgta 541 cccccgtggt ctttgaataa agtctgagtg ggcggc //
Select your sequence and copy it to your clipboard.
Go to the ORF finder at NCBI
Paste your sequence into the query box and hit OrfFind (the button is oddly placed above the data entry box). You will get something that looks like this:My translated sequence is in the +2 frame and is 429 bases long.
Answer these questions for your sequence:
- What is shown in blue/green?
- How long is the translated portion?
- Are there any untranslated portions?
- Which frame is translated?
Next, take your sequence and go to nucleotide blast (blastn) at NCBI.
Copy your sequence into the query box. Select "human genomic + transcript in the database block.
Click blast.
When the results are shown, select "human genome view" from the other views option.
This will take you to a screen which will show you which chromosome your gene is on.
My protein, hemoglobin a is on chromosome 16.
What chromosome is your gene on?
If you click on the chromosome, you will go to the map viewer. Find a region that shows high identity with a red box in the gene seq map (3rd line) and click on the blue text. Select "Sequence Viewer".
Selecting sequence viewer will bring up something that looks like this:
Dark green boxes with arrows are exons, light green boxes are untranslated regions of the mRNA and non-boxed green lines are introns. I can see that Hemoglobin a has two introns and 3 exons. I can also see the 5' and 3' UTR's. If I were to scroll to the left on this sequence, I should be able to find the promoter sequence.
- How many exons is the gene composed of?
- How many introns does it contain?
- Can you find your 5'UTR and your 3'UTR?
- Where would you expect to find your start and stop codons?







No comments:
Post a Comment